Building a Multimodal Mood & Stress Forecasting Pipeline — with Explainable AI (XAI)
Wearables can capture rich physiological signals minute-by-minute — but turning that into something interpretable and useful is a real engineering + ML challenge.
I recently completed an end-to-end project: Multimodal Mood & Stress Forecasting with XAI, where the goal is to forecast future stress (3-class ordinal) and mood/feeling valence (3-class) from wearable physiology + timestamped self-reports.
What I built (in plain English)
This repository contains a full pipeline that:
- Loads & cleans minute-level physiological data + self-report labels
- Aligns them by participant and timestamp
- Builds sliding windows (120 minutes window, 5 minutes stride) and creates robust time-series features
- Trains multiple models across multiple forecasting horizons (2h, 6h, 24h)
- Generates explanations so the model outputs aren’t a “black box” (SHAP + Integrated Gradients)
The dataset (high level)
- Physiology: 28 participants, minute-level signals (e.g., HR, HRV RMSSD, EDA, respiration, temperature, steps/accel, sleep/posture/wearing detection, missing reasons), ~901,440 rows (Mar 2024–May 2025).
- Self-reports: 4,668 timestamped entries (feeling, stressLevel, eventType). Duplicates are retained (~71%) for transparency, and can be optionally deduplicated depending on analysis goals.
This setup reflects a common real-world problem: labels are sparse while sensors are dense — so the pipeline includes per-horizon window-quality rules designed to handle sparsity more realistically.
Evangelista, E., Nazir, A., Bukhari, S. M. S., Dahmani, N., Tbaishat, D., & Sharma, R. (2025). Zayed University Physiological Wellness Dataset 2025 (ZU-PWD ’25): Longitudinal Multimodal Physiological and Behavioral Signals from Medical-Grade Wearables. Zayed University.
Models + Explainability (XAI)
I implemented both classic baselines and sequence models:
- Baselines: logistic regression, ordinal regression, LightGBM, XGBoost
- Sequence models: BiLSTM, BiGRU, TCN, Transformer encoder (dual heads for stress + feeling)
For explainability:
- SHAP for tree/linear models
- Integrated Gradients (Captum) for deep sequence models
Artifacts are exported under
artifacts/explanations(including per-participant summaries).
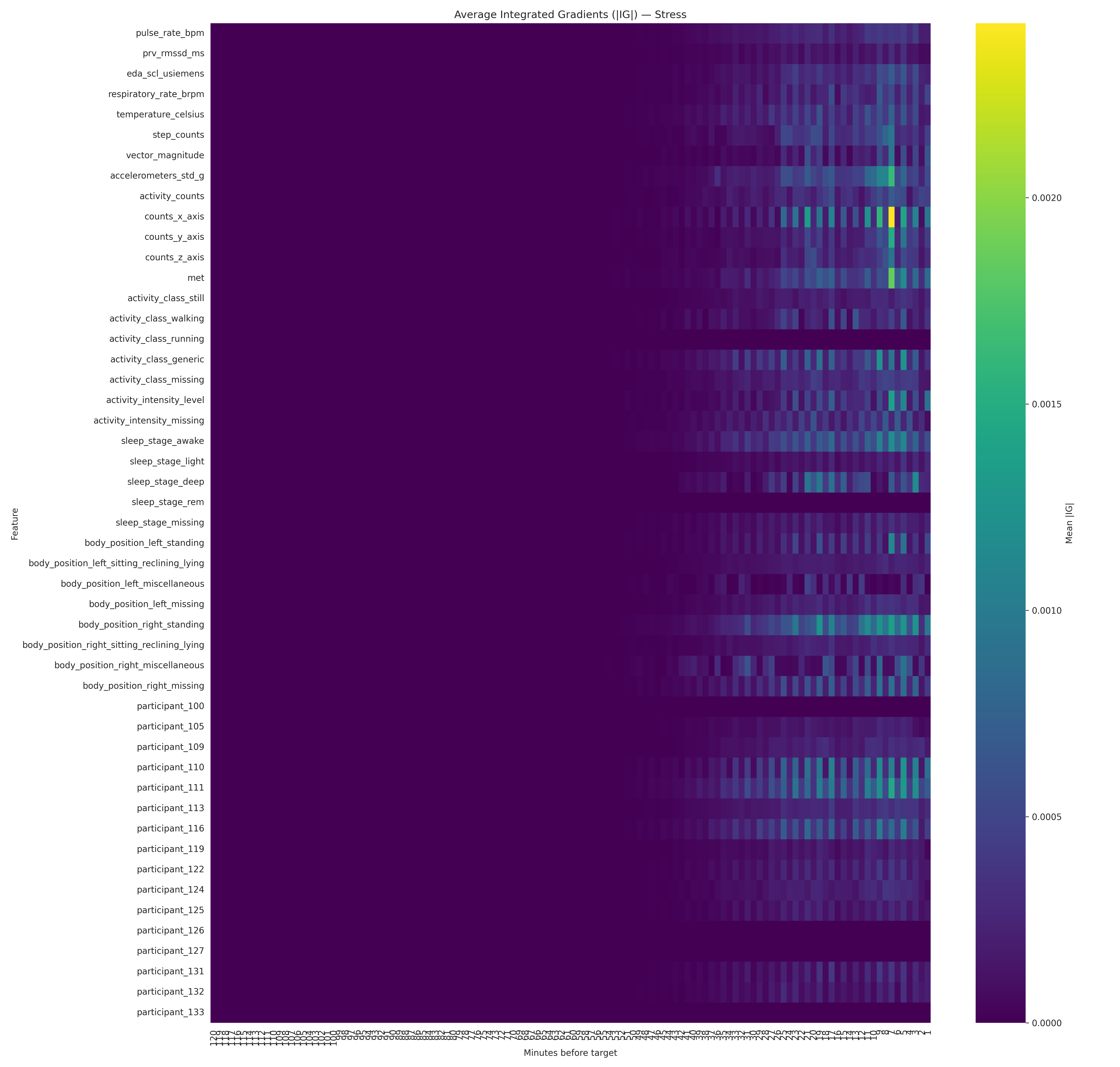
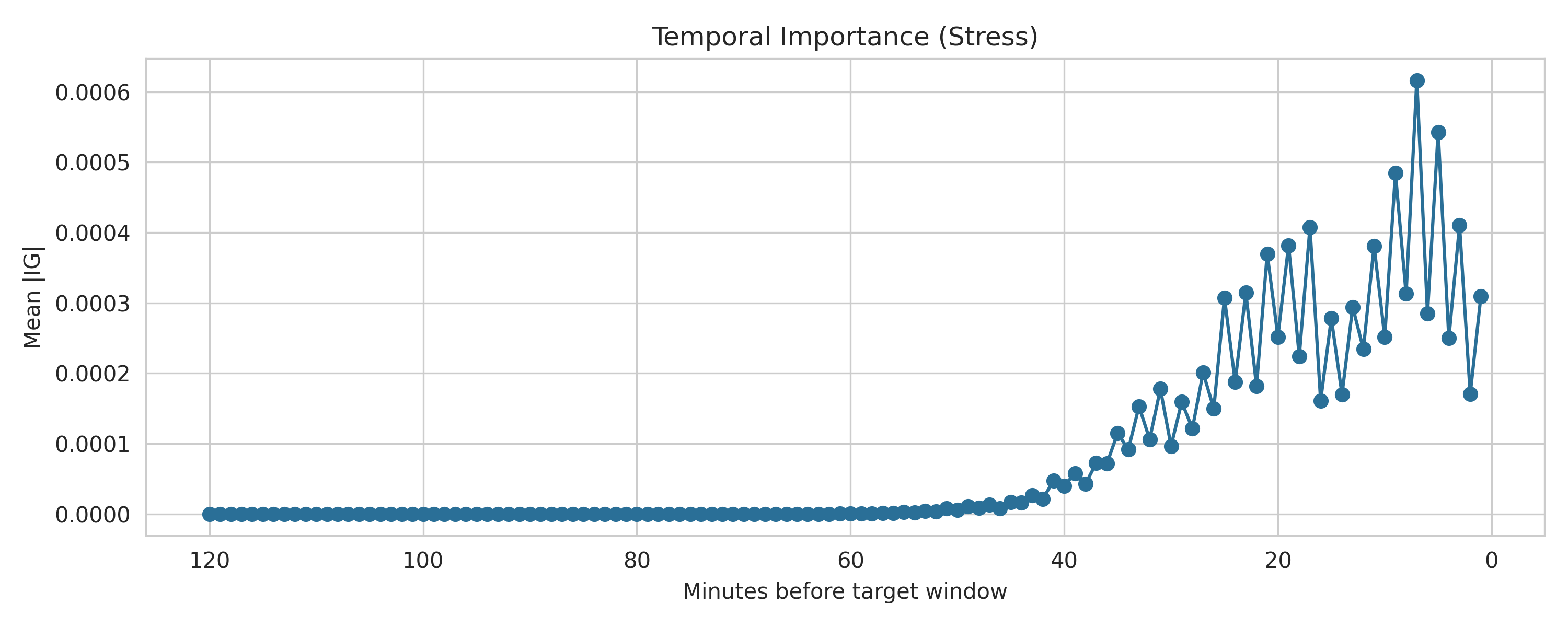
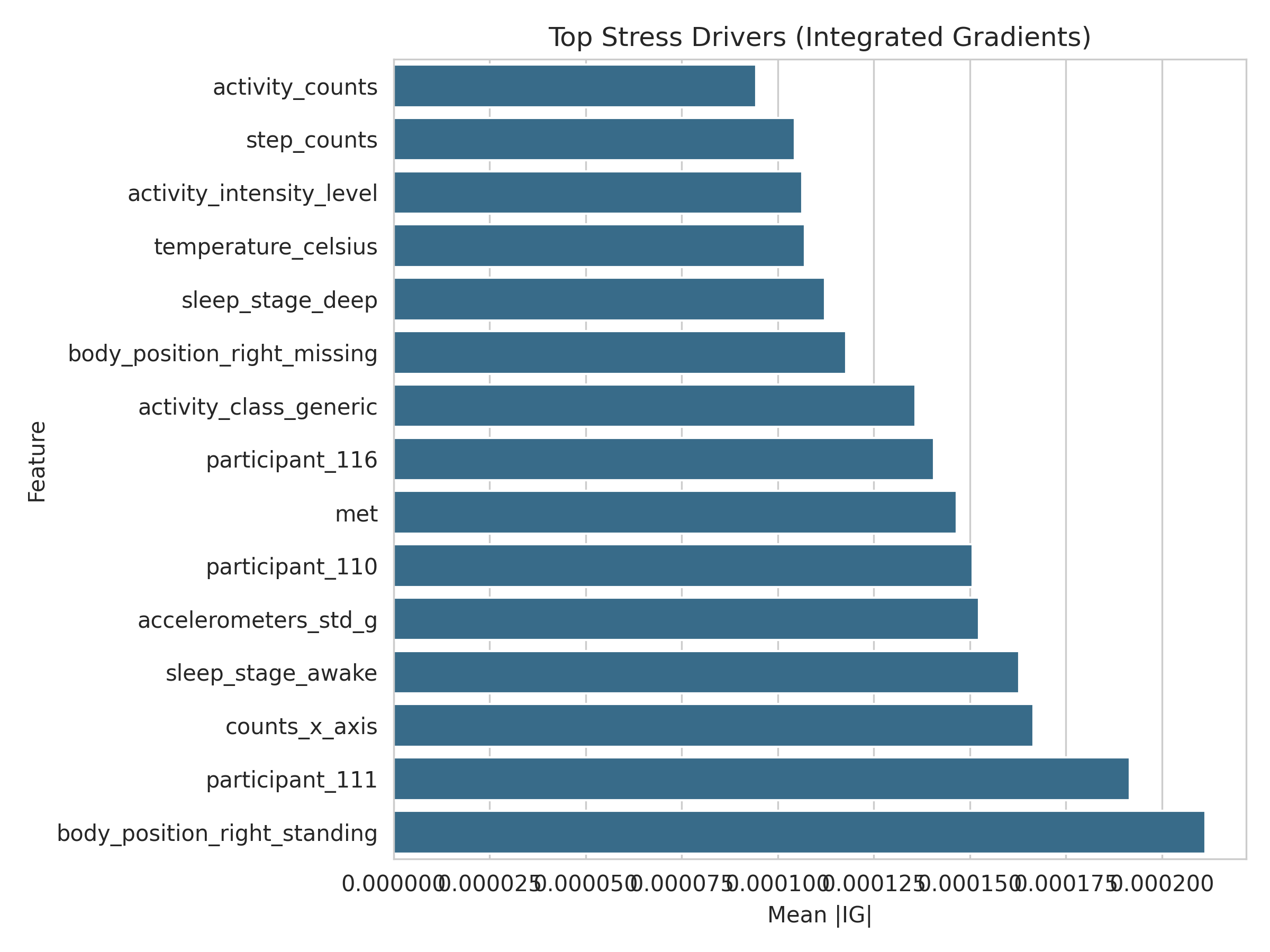
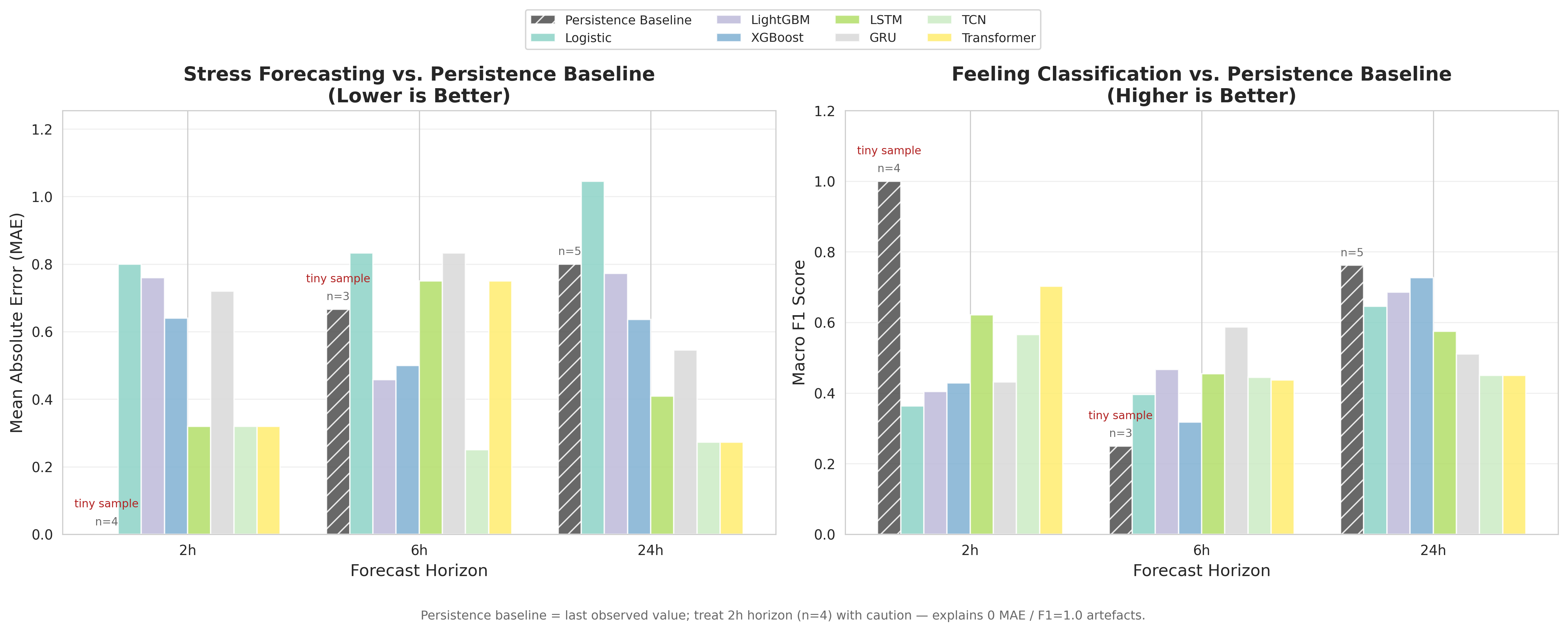
Results snapshot (with an important caveat)
A quick summary across horizons (see full tables in artifacts/REPORT.md and training_summary.json):
- 2h: best Feeling macro-F1 ≈ 0.70 (Transformer), best Stress MAE ≈ 0.32 (LSTM/TCN/Transformer)
- 6h: best Feeling macro-F1 ≈ 0.59 (GRU), best Stress MAE ≈ 0.25 (TCN)
- 24h: best Feeling macro-F1 ≈ 0.73 (XGBoost), best Stress MAE ≈ 0.27 (TCN/Transformer)
Caveat: the test folds can be very small (≤25 windows), so variance is expected — I explicitly note this in the repo to avoid over-claiming performance.
Why I’m sharing this
I wanted a repo that’s not just “train a model,” but something closer to what you’d need in practice:
- handling messy physiological data
- building windows + quality filters
- comparing strong baselines vs deep models
- producing explanations that you can actually inspect and discuss
If you’re working on wearables, time-series forecasting, multimodal learning, or explainable ML, I’d love your feedback.
GitHub repo
https://github.com/r3kind1e/Multimodal-Mood-Stress-Forecasting-with-Explainable-AI
If you check it out, the repo includes “view results without rerunning” entry points (report + artifacts + notebooks).